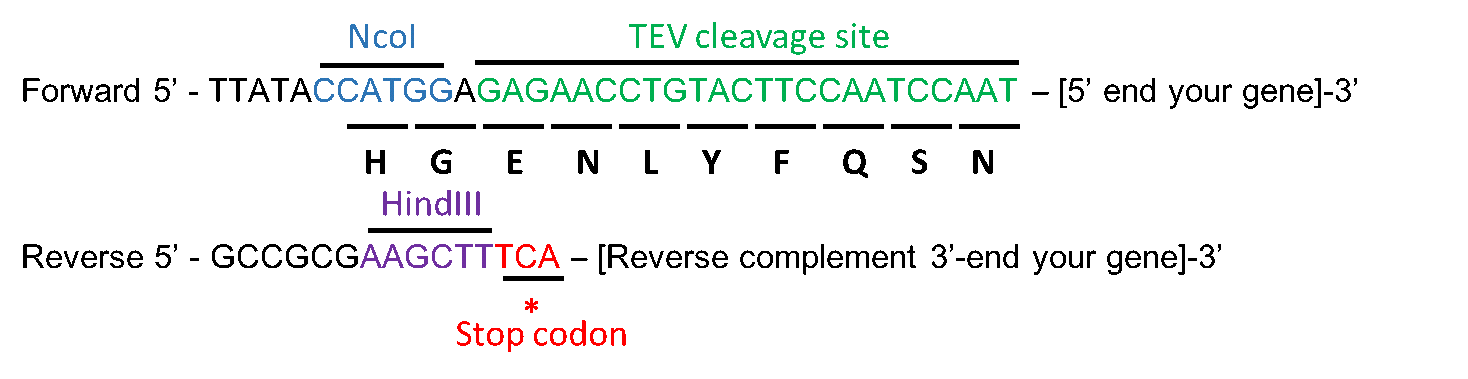
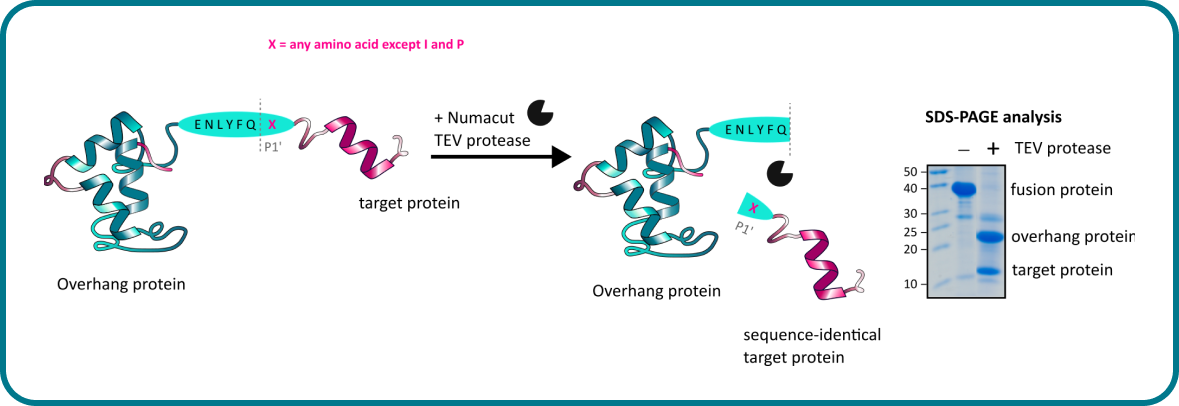
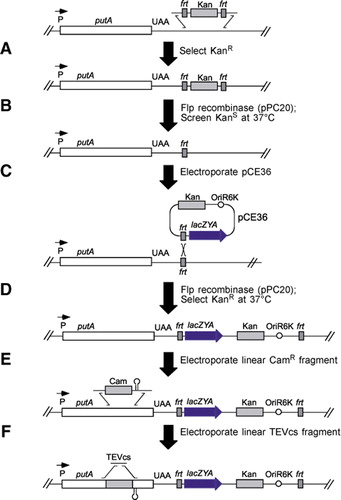
Recombinant Production of the Amino Terminal Cytoplasmic Region of Dengue Virus Non-Structural Protein 4A for Structural Studies | PLOS ONE

TEV protease cleavage of bioGATA-2 bound to streptavidin beads. (A)... | Download Scientific Diagram

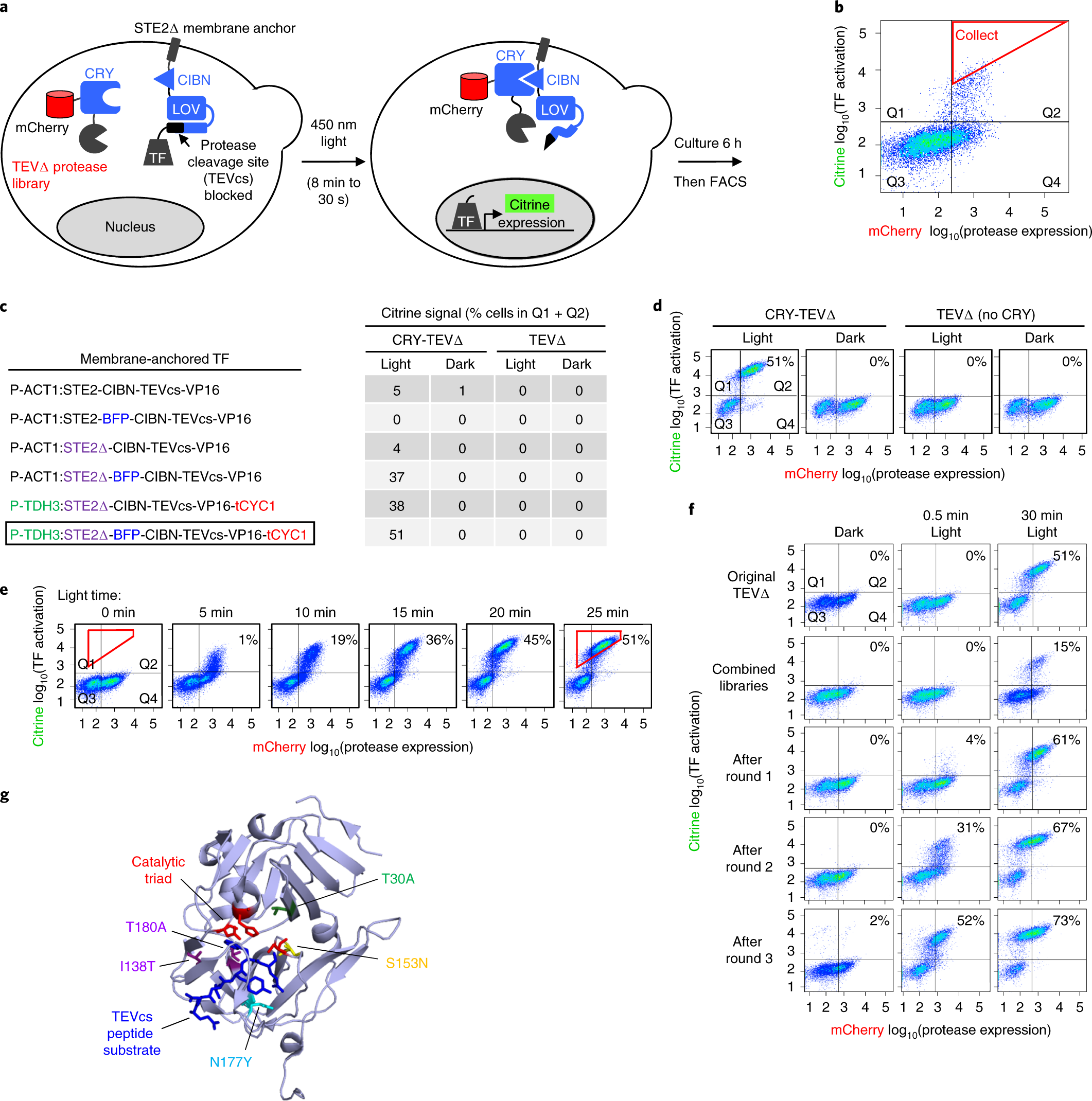
Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries | PNAS
Tobacco Etch Virus (TEV) protease protein fusions and immobilization... | Download Scientific Diagram

SDS-PAGE analysis of TEV protease cleavage efficiency. (A) 0.94 mg and... | Download Scientific Diagram

YESS 2.0, a Tunable Platform for Enzyme Evolution, Yields Highly Active TEV Protease Variants | ACS Synthetic Biology
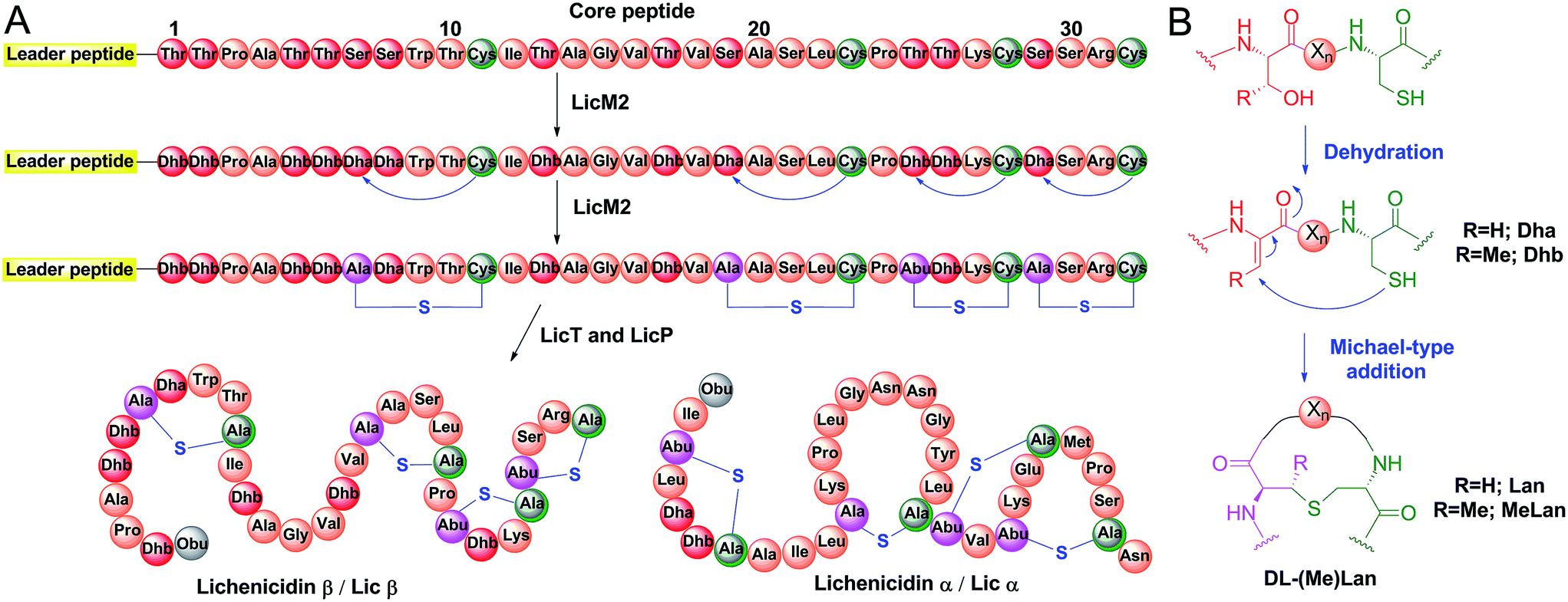
Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage - Chemical Science (RSC Publishing) DOI:10.1039/C5SC02329G
Highly efficient soluble expression, purification and characterization of recombinant Aβ42 from Escherichia coli

Three-Amino Acid Spacing of Presenilin Endoproteolysis Suggests a General Stepwise Cleavage of γ-Secretase-Mediated Intramembrane Proteolysis | Journal of Neuroscience

A fully automated procedure for the parallel, multidimensional purification and nucleotide loading of the human GTPases KRas, Rac1 and RalB. - Abstract - Europe PMC